Burmese grape (Baccaurea ramiflora Lour.) belongs to the family of Phyllanthaceae and is a fruit crop that originated in northeastern India to southern China and Peninsula Malaysia (POWO 2022). As a nutritional fruit, the species is not only rich in vitamin C and minerals, but also produces antioxidant compounds (Padayatty et al. 2003; Sundriyal and Sundriyal 2004). With a blend of sweet and sour, the pulp could be utilized in the food industry due to its economic efficiency and commercial importance. In the Mekong Delta, an indigenous variety of well-known Burmese grapes are Ha Chau, Green, Yellow, Red, and Xiem cultivars.
Ha Chau Burmese grape has light yellow color, thin skin, long fruit shapes, fruit has 3-4 segments, sweet and sour taste and specific aroma. Ha Chau Burmese grape was selected from indigenous cultivars by gardeners in Phong Dien district and developed in neighboring areas (Tran and Le 2012). The cultivar was recognized by the Intellectual Property Office of Vietnam as a specialty of Phong Dien district, Can Tho city in 2006 (Nguyen et al. 2018). Ha Chau Burmese grape has become a typical plant in Phong Dien district, Can Tho city, Vietnam and their plantation is the main income source for many horticulturists.
The propagation of Ha Chau Burmese grape is often through the intercrossing of selected plants with traits of interest. Nevertheless, long-term intraspecific hybridization could lead to inbreeding depression and thus, diminishing genetic background. On the other hand, cultivar identification with traditional techniques would be restricted due to the similarity in morphological features (Tran and Le 2012; Kodsara et al. 2021). Two popular molecular identification methods are AFLP (amplified fragment length polymorphism) and RAPD (random amplification of polymorphic DNA). However, each method has its limitations, such as the high cost of the AFLP technique and the accuracy of the RAPD technique (Kostamo et al. 2013). Therefore, the development of DNA based method for Burmese grape cultivar identification is of great potential.
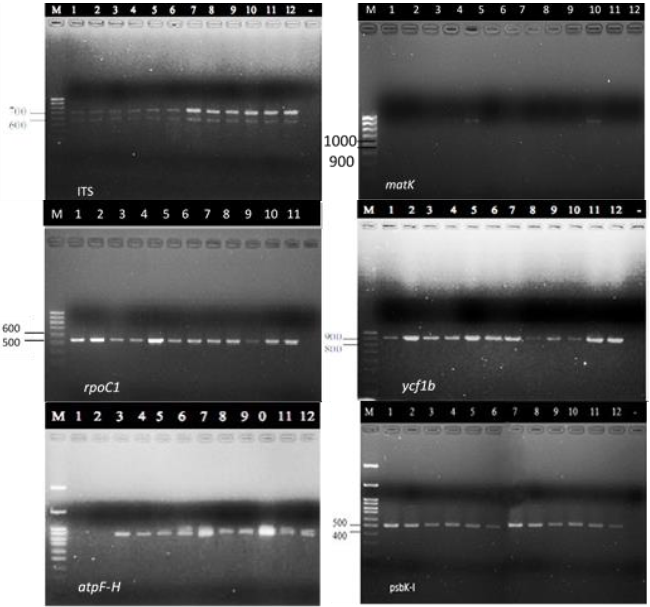
Species-specific DNA barcodes have been an effective and reliable approach for genetic diversity evaluation as well as cultivar identification. This method provides a powerful tool for distinguishing various organisms, including animals, plants, and microbes at genus or species levels due to the comparison of a short and standardized genetic region (DeSalle and Goldstein 2019). Kodsara et al. (2021) reported that plastid loci and rpoC1 could be utilized for discrimination of nine Phyllanthus species. The combination of barcoding and phylogenetic analysis indicated that P. acidus was the most genetically distinct from the rest of the analyzed species in the group. At the same time, this study also confirmed the close relationship between two pairs of species P. emblica - P. urinaria and P. emblica - P. reticulatus. Some DNA barcodes commonly applied in plant taxonomy are known as matK, psbA-trnH, rpoB , rpoC1 , psbK- I , atpF -H, trnL - trnF (Kress 2017) . Therefore, this study aimed to characte rize genetic diversity among the five cultivars of Burmese grape in the Mekong Delta and to distinguish Ha Chau to other cultivars based on DN A barcodes.
Data analysis Target sequences of Burmese grapes amplified were analyzed with corresponding data in the GenBank database to investigate barcode sequences. Sequences were aligned by Clustal W algorithm in Bioedit program. The consensus sequence for each cultivar was created from individual sequences, and variable sites were enumerated. Phylogenetic tree was constructed by Maximum Likelihood method in MEGA X software (Kumar et al. 2018) with Kimura-2-Parameter (K2P) model and 1000 bootstrap replicates. Other options were followed as default setting.
Data from this finding illustrated that the combination of five loci, namely matK, rbcL, trnL-F, psbA-trnH, and ITS indicated accurate species resolution while the single rbcL was less effective. Vu et al. (2020) also suggested that combination of plastid DNA barcodes increased the species identification in Vietnamese Paphiopedilum species. At the genome level, phylogenetic tree generated by 14 chloroplast genomes elucidated that B. ramiflora showed a closed genetic relationship with Phyllanthus, Glochidion, and Flueggea species (Hu et al. 2021).
In conclusion, the Burmese grape cultivars had genetic diversity based on the combination rcbL, rpoC1, ycf1b, and psbK-I sequences. This finding supports the idea that the sequence region at atpF-H could be used to distinguish Ha Chau Burmese grape from other cultivars in the Mekong Delta. Further studies on larger sample sizes and survey sites or the remaining regions should be carried out to investigate the appropriate sequence for identifying Ha Chau cultivar among others.
Nguồn bài viết: DOI: 10.13057/biodiv/d230726 B I OD I V E R SI T A S Volume 23, Number 7, July 2022 Pages: 3513-3520
Ngày đăng: 26-05-2025
Tác giả: Nguyễn Thị Bích Như